
The role of CRISPR in microbiome engineering [Podcast]
The Role of CRISPR in Scientific Breakthroughs in Microbiome Engineering
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) was first discovered in the genome of marine bacteria. When faced with a viral threat, bacterial cells developed an immune response by capturing and copying DNA fragments of viruses. This allowed bacteria to recognize subsequent attacks and cleave the viral DNA to stop the viral infection. It was also discovered that the Cas enzyme was responsible for DNA cleavage. This defense mechanism was later leveraged by Doudna and Charpentier, who could target a specific DNA sequence and isolate it using the CRISPR-Cas9 system(1).
Over the last decade, CRISPR-Cas9 has proven immensely valuable in drug discovery and drug manufacturing. Using a synthetic guide RNA (gRNA), scientists can target a specific DNA sequence and employ Cas9 for cutting it. Subsequently, the host repair machinery tries to repair the DNA through non-homologous end joining (NHEJ), leading to random mutations that alter gene function. Using this mechanism, scientists could now silence genes to elucidate their roles in disease phenotype, which can benefit target discovery. Furthermore, scientists can also use CRISPR-Cas9 to unravel mechanisms of drug resistance by identifying the set of genes associated with immune system evasion.
Today, however, we will explore a different application of CRISPR: microbiome engineering. Brought to you by Molecular Devices, the recent episode of the Drug Target Reviews Podcast discusses the applications of CRISPR in microbiome engineering and how it can overcome the bottleneck of human microbiome research.
Join leading experts Dr Jakob Haaber, Vice President & Head of Delivery Technologies, SNIPR Biome and Dr Richard Fox, Co-Founder, CEO and CTO of Infinome Biosciences, as they discuss the wide range of uses for CRISPR, including for therapeutics and biomanufacturing.
How CRISPR-Cas9 helps explore the role of human microbiome in diseases
It is now well known that the human body contains trillions of microorganisms, thereby outnumbering human cells. Collectively called the microbiome, these microorganisms form a symbiosis with the body by regulating the extracellular environment and protecting cells against pathogens. So, it is no surprise that disruptions in the microbiome are closely associated with numerous diseases, from diabetes and obesity to cancer.
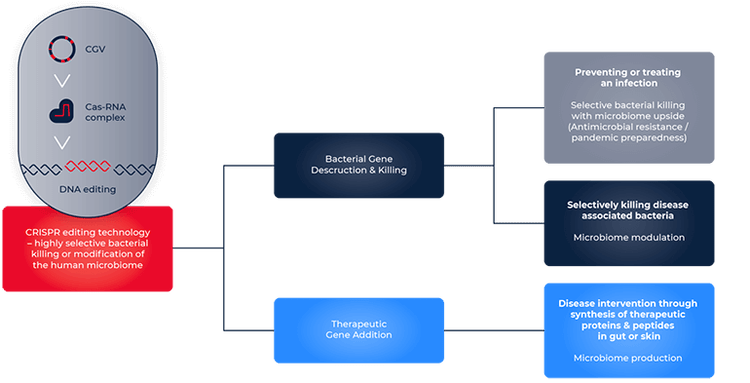
One way CRISPR contributes to drug discovery is its implementation to alleviate the disruptions in the microbiome. This is achieved by engineering CRISPR-Cas9 systems to cut specific bacterial DNA sequences to eliminate pathogenic bacteria. SNIPR Biome is one of the companies actively working towards this goal. Dr. Jakob Haaber, Vice President and head of CRISPR and Delivery Technologies at SNIPR Biome, describes the application of CRISPR to pathogenic E.coli elimination: “We target them using our CRISPR that we have programmed to kill E.coli. Currently, the stand out of care for treating bacterial infections is with antibiotics. But there is an increased occurrence of antibiotic resistance, rendering antibiotics ineffective. The advantage of CRISPR is that it does not distinguish between antibiotic sensitive- and antibiotic-resistant bacteria''. He also suggests that CRISPR can specifically target pathogenic bacteria and spare the beneficial ones integral to the healthy gut microbiome, as opposed to antibiotics with detrimental effects on healthy bacteria alongside pathogens.
Advantages of CRISPR and recent developments
According to Richard Fox, co-founder, CEO, and CTO of Infinome Biosciences, the main strength of CRISPR comes from its ability to scale up genome editing. “Instead of random mutations on a small set of target genes, we can now precisely edit an entire pathway or genome, introducing hundreds of thousands of changes.”
Large-scale genome editing gives rise to commercially available cell libraries that researchers can integrate into a high-throughput phenotypic screening. These libraries eliminate the need to manually knockout a large number of genes for phenotypic profiling and propose a solution for the acceleration of drug discovery studies.
Another advantage of CRISPR technologies is the improved specificity. Microbiome gene editing is one of the fields reaping the benefits of specificity. CRISPR systems can be programmed to cut specific bacterial DNA or even remove genetic circuits within bacterial cell walls without killing the cell. Taken together, these advancements drive novel gene therapies for gut microbiota deviations.
In the meantime, CRISPR systems have enabled the manufacturing of smaller genetic constructs for easier packaging into delivery vehicles. Thus, the delivery vehicle can carry multiple components in addition to CRISPR, making up a multifunctional gene-based therapeutic that accomplishes a lot more than just cutting or inserting DNA.
Acceleration in the manufacturing of CRISPR technologies
The gap between synthetic biology and bioengineering is often caused by issues regarding time and cost. Bringing a biomanufacturing solution to market requires large entities with well-built infrastructure, robust automation, lots of people, instrumentation, informatics, and a lot of capital. So, it is not surprising that the development of biologics can take 5-10 years and many millions of dollars. This is quite a risky investment, so pharmaceutical companies often hesitate to partake in it, leaving territories with huge therapeutic potential unexplored.
The setup of the CRISPR-enabled genome editing system is one of the bottlenecks in workflows. The design of large-scale genome engineering systems is quite laborious because one may need to produce minute amounts of donor sequence to target and precisely edit 10,000 loci into a whole genome.
One of the co-founders of Infinome Biosciences, Andrew Garst, came up with a key innovation by pairing the guide sequence that directs the cut with the donor sequence that mediates the repair, thereby reducing wide-genome editing to just a few clicks. Richard Fox believes “such automated design and build systems can create edited cell libraries in less than a week, which is the fraction of the effort it used to take”.
Steps of microbial gene editing
The first step in gene editing for the gut microbiome is the in vitro validation of the CRISPR system. The specificity of the CRISPR editing is tested against a set of bacterial panels representative of the gut microbiome. The aim is to ensure that the CRISPR system targets a predetermined subspecies of bacteria and that the beneficial bacteria are exempted from CRISPR-induced gene editing
Then, the validation carries on to the preclinical and clinical studies. Using genome sequencing techniques, researchers demonstrate that gene editing does not perturb the microbiome in a harmful way. Of course, off-target effects could inevitably occur, as revealed by whole-genome sequencing. The course of action is to monitor the rate of occurrence and ensure that these effects are trivial and do not interfere with gene editing.
Sequencing and phenotypic profiling reveal the set of edits that a genome carries as well as the prevailing strains in a bacterial population. To further validate the success of targeted genome editing, one can subject the bacterial population to environmental stressors and monitor their behavior. This ensures that the gene editing confers the bacteria to the desired properties, for instance the ability to grow under an environmental stress such as hypoxia.

With accelerated CRISPR workflows, researchers can elicit the set of beneficial gene edits amongst hundreds of thousands of gene knockouts.
CRISPR systems also unlock the flexibility to enrich or deplete the same bacterial strain depending on the specific research need. For example, researchers at Infinome implemented CRISPR to engineer E.coli strains to scale up lysine production, a critical amino acid used as a food additive. On the other hand, SNIPR used the same system to eradicate detrimental E.coli strains from the gut of hematological cancer patients to prevent bloodstream infections.
Looking ahead to the future of CRISPR
As mentioned before, CRISPR empowers the fight against pathogenic bacteria. It has been proven several times that conventional antibiotic treatments disrupt the gut microbiota by targeting both harmful and beneficial bacteria. CRISPR can help enhance the specificity of bacteria to maintain the balance of the human gut microbiota.
Another exciting prospect driven by CRISPR is the ability to engineer bacterial strains with therapeutic potential. This can be used to add genetic functions to the microbiome through engineered bacteria, which can express a specific enzyme or metabolite that the body previously lacked.
Lastly, the successful implementation of CRISPR in biomedical and bio-industrial engineering relies on the ability to make combinatorial edits, also known as “DNA shuffling”. Especially in large microbial systems, the ability to introduce multiple edits to reduce the number of screening rounds will be paramount to implement bio-industrial production.
Molecular Devices helps scientists by providing cutting-edge technologies that accelerate the immediate goal of CRISPR-Cas9 genome editing. An accurate library screening and selection of the CRISPR-edited hits is key within microbiome engineering workflows, allowing for faster availability of the biomanufacturing product to the market.
- Doudna, Jennifer A., and Emmanuelle Charpentier. "The new frontier of genome engineering with CRISPR-Cas9." Science 346.6213 (2014): 1258096.
CRISPR在微生物组工程科学突破中的作用
CRISPR首次在海洋细菌的基因组中发现,当面临病毒威胁时,细菌细胞通过捕获和复制病毒的DNA片段来产生免疫反应,这使得细菌能够识别随后的攻击并切割病毒DNA以阻止病毒感染。另外,还发现Cas酶负责切割DNA,这种防御机制后来被Doudna和Charpentier利用,他们可以靶向目标DNA序列,并使用CRISPR-Cas9系统将其分离(1)。
在过去的十年中,CRISPR-Cas9已被证明在药物发现和药物制造中具有巨大的价值。使用合成向导RNA(gRNA),科学家可以靶向目标DNA序列并使用Cas9进行切割。随后,宿主修复机制试图通过非同源末端连接(NHEJ)修复DNA,导致改变基因功能的随机突变。利用这种机制,科学家们现在可以使基因沉默来阐明它们在疾病表型中的作用,这有利于发现目标。此外,科学家还可以利用CRISPR-Cas9,通过识别与免疫系统逃避相关的一组基因来揭示耐药机制。
然而,今天,我们将探索CRISPR的不同应用:微生物组工程。本期内容我们来讨论CRISPR在微生物组工程中的应用以及它如何克服人类微生物组研究的瓶颈。
本次嘉宾:Jakob Haaber博士,副总裁兼交付技术主管SNIPR Biome和Infinome Biosciences联合创始人、CEO 兼CTO Richard Fox博士一起探讨CRISPR的广泛用途,包括治疗和生物制造。
CRISPR-Cas9如何帮助探索人类微生物组在疾病中的作用
众所周知,人体含有数万亿种微生物,在数量上超过了人体细胞。这些微生物统称为微生物组,通过调节细胞外环境和保护细胞免受病原体侵害,与人体形成共生关系。因此,从糖尿病、肥胖到癌症,微生物组的破坏与许多疾病密切相关也就不足为奇了。

通过设计CRISPR-Cas9系统来切割特定的细菌DNA序列以消除致病细菌,这种特异性使CRISPR技术减轻对整体微生物组的破坏,实现药物发现进程的加速。。SNIPR Biome是积极致力于实现这一目标的公司之一,SNIPR Biome副总裁兼CRISPR负责人和交付技术主管Jakob Haaber博士描述了CRISPR在致病性大肠杆菌消除中的应用:“我们使用CRISPR杀死大肠杆菌。目前,治疗细菌感染最常用的就是抗生素。但抗生素耐药性的发生率增加,进而使抗生素无效。CRISPR的优点是它不区分抗生素敏感和抗生素耐药细菌。他还建议,CRISPR可以专门针对致病细菌,而不影响健康肠道微生物组中不可或缺的有益细菌,而不是像抗生素一样,对健康细菌和病原体都有影响。
CRISPR的优势和近年发展

Infinome Biosciences的联合创始人、CEO兼CTO Richard Fox表示,CRISPR的主要优势在于其扩大基因组编辑的能力。“我们现在可以精确地编辑整个通路或基因组,而不是一小部分目标基因上的随机突变,从而引入数十万个变化。
大规模的基因组编辑产生可商用的细胞库,研究人员可以将其整合到高通量表型筛选中。这些库消除了手动敲除大量基因进行表型分析的需要,并为加速药物发现研究提出了解决方案。
CRISPR技术的另一个优点是提高了特异性,微生物组基因编辑是受益于特异性的领域之一。CRISPR系统可以被编程为切割特定的细菌DNA,甚至在不杀死细胞的情况下去除细菌细胞壁内的基因回路。综上所述,这些进步推动了肠道微生物群偏差的新基因疗法。
与此同时,CRISPR系统已经能够制造出更小的基因构建体,以便更容易地装配到递送载体中。因此,除了CRISPR之外,递送载体还可以携带多种成分,构成一种多功能的基于基因的疗法,不仅仅是切割或插入DNA。
加速CRISPR技术的制造
合成生物学和生物工程之间的差距通常是由时间和成本问题引起的。将生物制造解决方案推向市场需要大型实体,这些实体具有完善的基础设施、强大的自动化、大量的人员、仪器、信息学和大量资金。因此,生物制剂的开发可能需要5-10年和数百万美元也就不足为奇了。这是一项风险相当大的投资,因此制药公司往往不愿参与其中,从而一些具有巨大治疗潜力的领域未得到开发。
支持CRISPR的基因组编辑系统的设置是工作流程的瓶颈之一。大规模基因组工程系统的设计非常费力,因为需要产生微量的供体序列来靶向并精确地将10,000个位点编辑到整个基因组。
Infinome Biosciences的联合创始人之一Andrew Garst提出了一项关键创新,将指导切割的引导序列与介导修复的供体序列配对,从而将广基因组编辑减少到只需点击几下。Richard Fox认为,“这种自动化设计和构建系统可以在不到一周的时间内创建编辑过的细胞库,这是过去所花费的努力的一小部分”。
微生物基因编辑步骤
肠道微生物组基因编辑的第一步是CRISPR系统的体外验证。CRISPR编辑的特异性是针对一组代表肠道微生物组的细菌进行测试的,以确保CRISPR系统针对预定的细菌亚种,并且使得有益细菌不受CRISPR诱导的基因编辑的影响。
然后,验证继续进行到临床前和临床研究。利用基因组测序技术,研究人员证明基因编辑不会以有害的方式扰乱微生物组。当然,脱靶效应不可避免地会发生,正如全基因组测序所揭示的那样。我们需要做是监测发生率,并确保这些影响是微不足道的,不会干扰基因编辑。
测序和表型分析揭示了基因组携带的一组编辑以及细菌种群中的主要菌株。为了进一步验证靶向基因组编辑的成功,可以将细菌种群置于环境压力源中并监测其行为。这确保了基因编辑赋予细菌所需的特性,例如在缺氧等环境压力下生长的能力。

通过加速的CRISPR工作流程,研究人员可以在数十万个基因敲除中得到一组有益的基因编辑。
CRISPR系统还可以根据特定的研究需求,灵活地富集或消耗相同的细菌菌株。例如,Infinome的研究人员应用CRISPR来设计大肠杆菌菌株,以扩大赖氨酸的生产,赖氨酸是一种用作食品添加剂的关键氨基酸。另一方面,SNIPR使用相同的系统从血液癌患者的肠道中根除有害的大肠杆菌菌株,以防止血液感染。
展望
如前所述,CRISPR能够对抗致病菌,已多次证明,传统的抗生素治疗通过作用有害和有益细菌来破坏肠道微生物群。CRISPR可以帮助增强细菌的特异性,以维持人类肠道微生物群的平衡。
另一个令人兴奋的前景是设计具有治疗潜力的细菌菌株的能力。这可用于通过工程细菌为微生物组添加遗传功能,这些细菌可以表达身体以前缺乏的特定酶或代谢物。
最后,CRISPR在生物医学和生物工业工程中的成功应用依赖于进行组合编辑的能力,也称为“DNA洗牌”。特别是在大型微生物系统中,引入多次编辑以减少筛选轮次的能力对于生物工业生产至关重要。
Molecular Devices通过提供前沿技术来帮助科学家加速CRISPR-Cas9基因组编辑的近期目标。准确的文库筛选和CRISPR编辑命中物的选择是微生物组工程工作流程的关键,使生物制造产品更快地推向市场。
1. Doudna, Jennifer A.和Emmanuelle Charpentier。“CRISPR-Cas9基因组工程的新前沿。科学346.6213(2014):1258096。